According to Howard Barry Schatz: "Dorothy Garrod’s Canaanite Natufians would have been living in Middle Eastern caves since 13,000 BCE, more than 3000 years before the Holocene period began (ca. 9700 BCE). Soon after it began, Natufians would have emerged from their caves to begin construction on Göbekli Tepe. "
The writings of 1st century historian Flavius Josephus describe Adam’s vision of the earth’s destruction by flood and flame. Josephus also implies that Enoch, the first holy man and scribe, carved the Razah D’Oraytah (Secret of Knowledge) into stone pillars to preserve this Knowledge for posterity after the prophesied flood. This article introduces evidence that the archaeological discovery of Göbekli Tepe on the Turkish plains of Haran, inadvertently uncovered these legendary “Pillars of Enoch.” This article endeavors to decipher the Enochian “Secret of Knowledge” that is carved into the pillars and architecture of Göbekli Tepe as history’s first theological and mathematical “treatise.” This challenges Assyriologist Samuel Noah Kramer’s assertion that “Sumerian men of letters developed no literary genre comparable in any way to a systematic treatise of their philosophical, cosmological, and theological concepts.” (Kramer, 1981: p. 79). This explains why Abraham’s father Terah longed to spend his final years in Haran, and why he named one of his sons Haran. What ties Enoch’s pillars to Göbekli Tepe are the revelations carved into its pillars and architecture. The one text capable of revealing these secrets is the Sefer Yetzirah: The Book of Creation, the only text attributed to Abraham. This text encrypts an ancient mathematical table known as the “231 Gates” (or “God Table”), which provides the underlying structure of the Abrahamic/Mosaic Word of God, 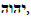 as well as Enoch’s Word of God,
as well as Enoch’s Word of God, 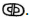 Both “Words” are rooted in the Luwian hieroglyphics for “God” and “Gate” that are carved into Göbekli Tepe’s “Blueprint of Creation.” This suggests that the pivot point between pre-history and history can be moved from the 4th millennium BCE invention of writing and mathematics to 10th millennium BCE Göbekli Tepe. These Göbekli Tepe revelations, including the DNA of its builders, enable us to trace civilization’s origins back to its Ethiopian founders.
Both “Words” are rooted in the Luwian hieroglyphics for “God” and “Gate” that are carved into Göbekli Tepe’s “Blueprint of Creation.” This suggests that the pivot point between pre-history and history can be moved from the 4th millennium BCE invention of writing and mathematics to 10th millennium BCE Göbekli Tepe. These Göbekli Tepe revelations, including the DNA of its builders, enable us to trace civilization’s origins back to its Ethiopian founders.